Unified Biomedical Knowledge Graph
Accepted Assertion File Formats
Objectives
This guide describes how to format a set assertions so that it can be added to the Unified Biomedical Knowledge Graph database.
The guide includes recommendations for optimizing and deepening the integration of a set of assertions into the UBKG to establish new relationships among entities and cross-references among ontologies.
Audience
This guide is intended for users who are subject-matter experts in what biomedical assertions they might want to represent (e.g., genes and their products) with a set of assertions, but not necessarily conversant with either ontological concepts or knowledge graphs.
Glossary
The Glossary page describes terms that this guide uses that are relevant to assertions or knowledge graphs.
Guiding Principles for Integrating a Set of Assertions
If the UBKG is to connect sets of assertions from different SABs, the sets of assertions should, as much as possible, represent entities and relationships similarly.
To the degree possible,
- Entities should be encoded with codes from published biomedical ontologies and vocabularies. For example, genes should be encoded in a standard vocabulary such as HGNC.
- Relationships should be represented with properties from the Relations Ontology.
- Codes for entities should be cross-referenced to UMLS CUIs (via the node_dbxrefs column, described below).
Format Options
The UBKG generation framework can work with files from a variety of sources, including:
OWL files
The generation framework can work with OWL files in serializations including:
- RDF/XML
- OWL/XML
- RDF
- Turtle
- OBO
- NCBO CSVs in GZIP archives
Files can be online or stored locally.
Custom data formats
Some data sources are available in custom formats, such as files in FTP sites. The UBKG development team can build custom scripts that extract and translate assertion content.
UBKG Edges/Nodes format
The rest of this document will discuss the UBKG Edges/Nodes format.
PubChem
Additional information to guide ingestion of PubChem information, a type of UBKG Edge/Node file, is here.
UBKG Edge/Node Format
Files in UBKG Edge/Node format describe the entities and relationships of a set of assertions that is to be integrated into the UBKG.
A file set of UBKG Edge/Node files consists of two Tab-Separated Variables (TSV) files:
- edges.tsv: Describes the triples comprising the ontology
- nodes.tsv: Describes metadata for entities
Compression
UBKG Edge/Node files can be compressed in GZIP:
- Each file should be in its own GZIP file.
- The filename of the GZIP should correspond to the file name of the contents.
In other words, if files are compressed, there should be files named
- edges.tsv.gz
- nodes.tsv.gz
Source Abbreviations (SABs)
A Source Abbreviation (SAB) is an uppercase acronym that identifies a set of assertions. The SAB is based on the UMLS SAB, which the UMLS uses to identify a vocabulary or domain. The UBKG extends the SAB identifier to include sets of assertions that are not in the UMLS.
The SAB is synonymous with an ontological namespace.
In general, a set of assertions can employ multiple SABs, of two types:
- Node SAB: The code for a subject or object node associates with the SAB that identifies the code’s steward. The steward maintain the code source. Examples of stewards include the organization that publishes an ontology to NCBO BioPortal or OBO.
- Edge SAB: A set of assertions associates with a SAB that may be different from the SABs of the nodes in the set.
The steward of a set of assertions is, in general, not the steward of all the codes involved in the assertions.
Example
Consider the following subset of assertions from the Phenotypic Quality Ontology (PATO). (Predicates have been translated from their labels in Relations Ontology.)
| subject | predicate | object |
|---|---|---|
| UBERON:0000457 | branching part of | UBERON:0001532 |
| PATO:0001776 | subClassOf | PATO:0001544 |
| CL:0000101 | capable of | GO:0050906 |
| PATO:0001894 | subClassOf | UBERON:0000061 |
This set contains three types of assertions:
- assertions involving only codes from PATO
- assertions involving codes from ontologies other than PATO (UBERON, CL, GO)
- assertions involving codes from PATO and other ontologies (e.g. PATO and UBERON)
The types of SABs in this set of assertions:
- SABs for the nodes: PATO, UBERON, CL, GO
- SAB for all the edges: PATO
In other words, PATO is the steward of a set of assertions that involve codes maintained by other stewards (e.g., UBERON).
When ingesting a set of assertions, it is necessary to define a preferred SAB to represent the steward of the set–e.g., an initiative, project, or institution.
edges.tsv
The edges file lists the triples (subject node - predicate - object node) that constitute a set of assertions.
Fields
| Field | Corresponding element in UBKG | Accepted formats | Examples |
|---|---|---|---|
| subject | Code node | OBO Principle 3 conforming (see note below) IRI for a concept in a published ontology | http://purl.obolibrary.org/obo/UBERON_0004086 |
| Code for the concept in the format SAB {space} code in ontology OR SAB {underscore} code in ontology. | UBERON 0004086, UBERON_004086 | ||
| predicate | relationships | For hierarchical relationships, the IRI http://www.w3.org/2000/01/rdf-schema#subClassOf OR the string “isa” | http://www.w3.org/2000/01/rdf-schema#subClassOf |
| For non-hierarchical relationships, an IRI for a relationship property in RO | http://purl.obolibrary.org/obo/RO_0002292 | ||
| Custom string that complies with neo4j relationship naming rules (see relationship labels below) | drinks milkshake of | ||
| object | Code node | same as for subject | |
| evidence_class (optional) | string | Statement specific to an SAB to classify evidence | -0.016084092 |
Requirements for nodes
The identifier (code) for a node must include the SAB for the code’s steward.
OBO Principle 3
The preferred form of identifier for a node is an IRI that conforms to Principle 3 for URI/Identifier Space. UBKG recognizes IRIs in this format implicitly.
An example of a IRI with the preferred format is: http://purl.obolibrary.org/obo/UBERON_0004086
In this example, UBERON is the SAB for the code.
For codes in a set of assertions that correspond to entities that have already been identified in a standard biomedical ontology, the IRI is preferred. For new codes that have not been already been encoded in an ontology, the identifier must include the SAB that represents the code’s steward.
Special characters and delimiters
UBKG uses complex rules for nodes for parsing SABs from nodes. The following characters are reserved for use as delimiters:
- space
- colon
- underscore
For best results in your nodes, use a colon, an underscore, or a space, to separate the SAB and code. For example, the following are all acceptable representations:
- UBERON:004086
- UBERON_0004086
- UBERON 004086
Do not use any of the reserved characters in node IDs.
Recommendations for edges
The preferred source of relationship (predicate) information is the Relations Ontology (RO). Reasons for this include:
- RO is a general reference for relationships, and is therefore likely already to have a standard relationship defined that is suitable.
- RO defines inverse relationships, especially those that may not be obvious.
It is possible, nevertheless, that RO does not contain a relationship that is specific enough for an assertion, so a custom relationship will be needed. When defining a custom relationship, we recommend that the label be short and concise. (This is easier said than done, of course. Defining concise relationships is the hard part of modelling assertions.)
Special characters and delimiters
The preferred format for a predicate is an IRI with format
http://purl.obolibrary.org/obo/RO_code
The format RO:code is accepted, but not preferred.
Relationship label format
The neo4 naming rules specify that relationship names:
- Contain only alphanumeric characters or the underscore.
- Start with a alphabetic character.
Although a relationship can include special characters or start with a number, Cypher queries that refer to the relationship must escape the relationship name with backticks. To avoid the need for backticks, relationships in the UBKG are reformatted as follows:
| Characters | Change | Example |
|---|---|---|
| hyphen | replaced with underscore | abc-def to abc_def |
| other non-alphanumeric characters | replaced with underscores | abc (def) to abc_def |
| leading numeric character | preceded with ‘REL_’ | 1abc to REL_1abc |
nodes.tsv
The nodes.tsv file provides metadata on entities.
Fields
| Field | Corresponding element in UBKG | Accepted formats | Examples |
|---|---|---|---|
| node_id | Code node | OBO Principle 3 conforming IRI for a concept in a published ontology | http://purl.obolibrary.org/obo/UBERON_0004086 |
| Code for the concept in the format SAB {space} code in ontology | UBERON 0004086 | ||
| node_label | Term node, Preferred Term (PT) relationship | Text string | Ventricles of hindbrain |
| node_definition (optional) | Definition node, DEF relationship | Text string | One of the system of communicating cavities in the brain …. |
| node_synonyms (optional) | Term node; Synonym (SYN) relationship | Pipe-delimited list of synonyms | See Example for synonyms below |
| node_dbxrefs (optional) | Cross-references | Pipe-delimited list of references to cross-referenced concepts. Each cross-reference should be in format SAB:code or UMLS:CUI | See Example for dbxrefs below |
| value (optional) | Numeric value | numeric | 20 |
| lowerbound (optional) | Lower bound of range for values | numeric | 5 |
| upperbound (optional) | Upper bound of range for values | numeric | 100 |
| unit (optional) | Unit of measure for value. This is currently not encoded to a concept. | string | mm |
Example for synonyms
region of ventricular system of brain|brain ventricles|cerebral ventricle
Example for dbxrefs
umls:c0007799|fma:78447
The UMLS cross-reference in the example is to a CUI; the fma cross-reference is to a code.
Requirements for nodes
The IRI requirements for nodes in the nodes file are identical to those for nodes in the edges file.
Requirements (business rules) for a set of assertions
- A node identified in edges.tsv must satisfy at least one of the following criteria:
- The node is defined in nodes.tsv. (Generally, this is for codes that are maintained by the steward of the set of assertions–i.e., the steward maintains both nodes and edges.)
- The node already exists in the UBKG. These nodes can be part of the UMLS or from a previously ingested set of assertions.
The UBKG generation framework will ignore nodes that do not satisfy at least one of these criteria.
- If a triple in edges.tsv refers to a node from a non-UMLS ontology, the non-UMLS ontology will need to be ingested into the UBKG first. For example, because the Mammalian Phenotype Ontology (MP) includes nodes from the Cell Ontology (CL), CL should be integrated into the UBKG before MP. This often improves the cross-referencing because the general ontologies have deeper external-referencing to UMLS and other OBO sources.
- This spreadsheet lists the SABs and example codes for the ontologies that are currently represented in the UBKG. It should be used as the reference for formatting existing source abbreviations (SAB) and their codes. In other words, if a SAB is already part of the UBKG, it should be sufficient to refer to the node by code.
For additional details regarding the UMLS SABs, please consult the UMLS reference.
- Some ontologies (including HGNC, GO, and HPC) include the SAB in codes (e.g., HGNC:9999) Nodes for concepts from these ontologies should be formatted as
: code-e.g., “HGNC HGNC:9999”. - The UBKG ingestion will reformat predicates so that strings are delimited with underscores.
- The UBKG ultimately requires that two nodes be linked with both a relationship and its inverse. However, in an edges file each relationship (predicate) should be represented only once (NOT with the original and inverse). The UBKG identifies and adds the inverse relationships using the RO as follows:
| predicate | form of inverse relationship | comment |
|---|---|---|
| IRI of relationship property in RO | Resolution via RO, using the IRI | |
| String that corresponds to the relation label (e.g., “has gene product”) | Resolution via RO, using relationship labels | not as precise as IRI |
| Custom string | Appends “inverse_” to the string | If the custom string actually corresponds to a relationship in RO with an ambiguous inverse, UBKG will create a new inverse relationship. |
| IRI of relationship property from ontology other than RO | none | The UBKG will ignore these relationships. |
Best Practices for cross-references
The items in the node_dbxref field of nodes.tsv establish cross-references. The degree to which an ontology integrates with the UBKG depends directly on cross-references: an ontology with nodes that cross-reference nodes in other ontologies are more likely to contribute to new relationships, while an ontology with few cross-references will essentially exist independently of the rest of the UBKG.
A best practice for cross-references is: the closer a cross-reference is to a UMLS CUI, the better.
The UBKG ingestion preferentially selects UMLS CUIs from the list in node_dbxref. It is not necessary to list both a UMLS CUI and a code from an ontology that is already in the UMLS. If it is not feasible to cross-reference a UMLS CUI, then try to cross-reference a code from a published, preferably OBO-compliant ontology.
Cross-references vs. isa
A cross-reference (dbxref) is not the same thing as an isa relationship.
isa
An isa assertion between two nodes indicates that the subject node relates to the object node hierarchically.
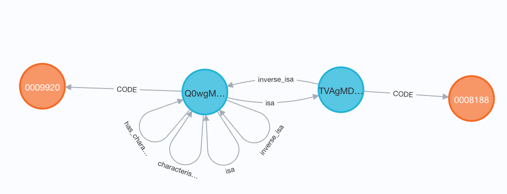
For example, the Mammalian Physiology Ontology (MP) asserts that MP:0009920 (abnormal t2 stage b cell morphology) isa MP:0008188 (abnormal transitional stage B cell morphology).
In the UBKG, the Code node for MP:0009920 has a path to the Code node for MP:0008188 through concepts that link via an isa relationship. Each Code node (orange) associates with a separate Concept node (blue). In other words, the UBKG considers the two Codes to represent different concepts.
dbxref
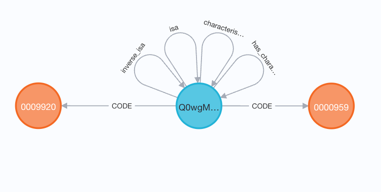
A dbxref establishes an equivalence between two nodes.
For example, MP cross-references MP 0009920 to CL:0000959 (T2 B cell).
In the UBKG, the Code nodes share the same Concept node. The two Code nodes represent the same Concept.
It is not necessary to link two concepts with both an isa relationship in the edge file and a node_dbxref in the node file.
Another way to think of this is:
- isa means “is a type of”
- dbxref means “is the same as”